Discussion
Summary
This time, the production of DNA origami, the design and experiment of the DNA circuit, and the implementation of the circuit using the origami and the implementation of the switch mechanism was confirmed. From this result, we can think that the idea we thought is feasible.
Evaluate our method
Mismatch
Besides,the experiment for the ON/OFF switch mechanism of fluorescent molecules is conducted by using mismatched base pairs. As a result, an ON / OFF mechanism is achieved, but it did not reach the point where the convergence value decreased from the initial concentration or the time of the OFF state was controlled. The current switch circuit delay mechanism using mismatched base pairs can be implemented with a seesaw gate and is expected to be achieved in future research. Also, as an alternative to mismatched base pairs,special self-bindings such as hairpin loops and bulge loops can be used to implement this idea.
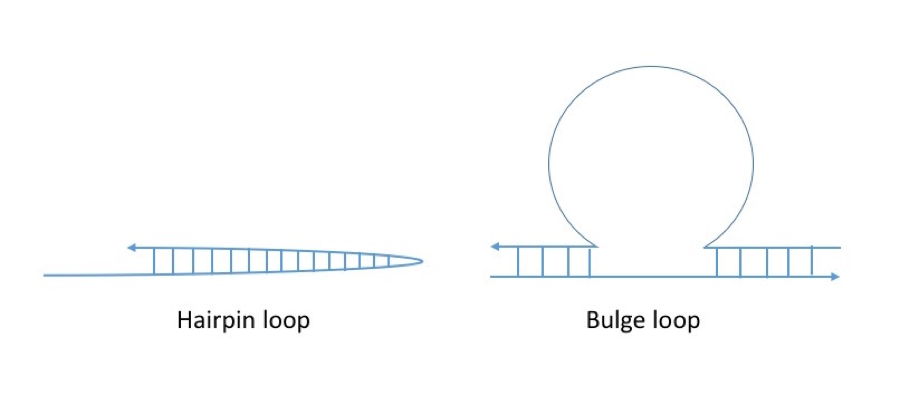
Figure1: Special self-bonding
DNA origami
This time, the design was based on origami, but some disadvantages arise from the use of origami. One of them is about the strength of DNA origami. As shown in the experiment, the planar structure of DNA origami is neatly sheet-like and rectangular as designed, but even though the origami is made, it can be confirmed that it is distorted or twisted. Since DNA is not as hard as chemical fibers, it is thought that deformed ones are born. Therefore, even though it can be designed to perform the strand displacement reaction on DNA origami, it seems difficult to create a clean form.
This is thought to be improved by creating an environment in which the planar structure of DNA origami becomes a sheet. Specifically, the method of fixing the four corners of the DNA origami rectangle with pins appears, but there are a lot of materials to consider such as what to fix and what plays the role of the pin Not realistic. Therefore, a more conceivable concrete plan is a method of incorporating DNA origami into a DNA origamithat has a nano-sized material such as a device and its mass. We think that this substance gives weight to light DNA (depending on the arrangement of the substance) and can suppress movements that lead to deformation. In addition, although the rectangle was adopted in this research, there is a possibility that it can be solved by changing the shape. In that case, it is necessary to consider the aspect depending on the size and weight of the material.
We think that the deformation of the origami can be suppressed by making the DNA origami as the foundation of several layers. Or there are plenty of ways to find it through trial and error.
Future
For display
We would like to propose the idea of developing “pixels”. In general, pixels used in a color display are expressed using three primary colors of R (red), G (green), and B (blue). In order to follow this, fluorescent molecules that emit three primary colors are attached to the planar structure created by DNA origami. To fluoresce the colors to be expressed based on the three primary colors, it is considered that various colors can be expressed by causing these fluorescent molecules to react while calculating the corresponding quenching molecules. However, the possibility of fret formation by attaching three fluorescent molecules on one plane must be considered here. However, DNA uses four bases, and there is a blueprint on the plate made with DNA origami. Therefore, we think that it is possible to calculate the distance at which frets do not occur and to design each of the fluorescent molecules attached thereto.
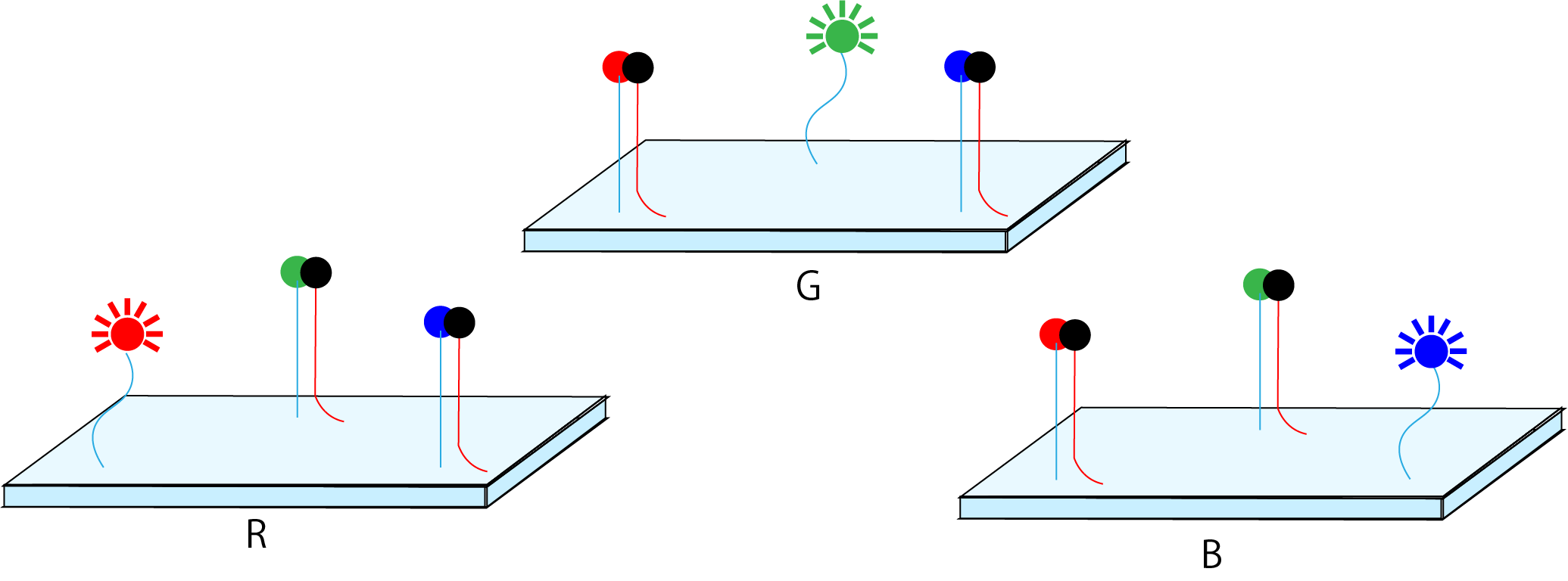
Figure2: Image element
ELSI
Most of the parts used in this project, including DNAo, consist of DNA. Therefore, it has a high affinity in vivo and has the potential to cause chemical reactions that cannot normally occur. In this project and experiment, it is not assumed to be used in vivo, but if a third party tries to abuse it, it can be injected into the body and used as a crime tool. In addition, if a DNA strand is introduced into a living body, the influence on the genes originally present in the living body must also be considered.
References
- Jinglin Fu, Minghui Liu, Yan Liu, Neal W. Woodbury, and Hao Yan, "Interenzyme Substrate Diffusion for an Enzyme Cascade Organizedon Spatially Addressable DNA Nanostructures", JACS, 2012.
- Qian Mei, Xixi Wei, Fengyu Su, Yan Liu, Cody Youngbull, Roger Johnson, Stuart Lindsay, Hao Yan, and Deirdre Meldrum, "Stability of DNA Origami Nanoarrays in Cell Lysate ", ACS Publication, 2011.
- NUPACK: http://www.nupack.org
- caDNAno: http://cadnano.org